Research directions
Molecular machines of the future
Our goal is to build ultra-miniaturized molecular devices and machines that can be combined into autonomously functioning systems ("robots") capable of executing user-defined tasks. At the moment we are primarily concerned with constructing molecular hardware for such purposes. To this end we investigate how to exploit the physical principles underlying the formation of natural molecular machines such as proteins for our purposes. Important principles are self-assembly, polymer folding and the fact that structures can be encoded in polymer building block sequences. Molecular self-assembly with DNA is an attractive route toward implementing these principles to create synthetic molecular machinery. DNA origami in particular enables building nanodevices that can already be employed for making new discoveries in biomolecular physics and protein science.

To achieve our goals, we work along a roadmap to systematically increase the structural and functional complexity of molecular structures that we can design and produce. This roadmap covers a wide range of subjects ranging from increasingly complex design puzzles for making components and mechanisms, fundamental physical self-assembly studies, biotechnological production of DNA molecules (our raw material), structural analysis using cryo transmission electron microscopy to computational modeling. Our tools and our researchers are by necessity multi-disciplinary.
Components and mechanisms through biomolecular self-assembly
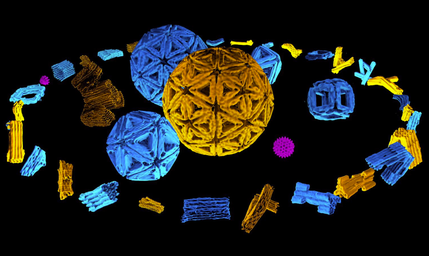
Designing the sequences of biomolecules to construct increasingly sophisticated molecular devices and machines is the key interest of this laboratory. Our method of choice is 'scaffolded DNA origami' and we are striving towards improving this method in order to engineer ever more complex devices that achieve more sophisticated functionalities. We use a comprehensive approach that includes various types of physical / chemical modelling, detailed experimental studies of assembly behaviour and structure, and research on mass production to push DNA origami toward practical applications. We have also embarked in designing proteins for creating hybrid DNA-protein structures.
Controlling and powering molecular motions
Creating molecular machines and motors that can work with efficiencies rivalling those of natural protein machines is an important unmet challenge. One obstacle toward realising such machines is the difficulty to build sufficiently sophisticated molecular structures. Another problem concerns incomplete understanding of the design principles for achieving the desired function. Exploring how to design increasingly sophisticated molecular structures is valuable, because it can yield an improved understanding of the links between sequence, shape, and function of biomolecules. Custom-designed macromolecular structures can also be employed as tools to support scientific discovery in various fields of research. Attempting to construct artificial molecular machines from biomolecules may enable an experimental test of models for the function principles of natural molecular machines. Artificial molecular machines and motors could help exploring how mechanical motion may be coupled to chemical reactions, and they may help to understand how increasingly complex enzymatic function can evolve from simple start mechanisms. But there is also a practical dimension: artificial molecular machines and motors could be used for example to drive chemical synthesis, to actively propel nanoscale drug delivery vehicles, to pump and separate molecules across barriers, or to package molecules into cargo components. We focus on DNA in particular as a programmable construction material to build nanometer-scale devices and mechanisms for applications in biomolecular physics, biological chemistry, and molecular medicine. 3D transmission electron microscopy and single molecule methods such as optical trapping and fluorescence microscopy are among our routine analysis tools.
(Sub-)Nanometer-precise device fabrication and validation

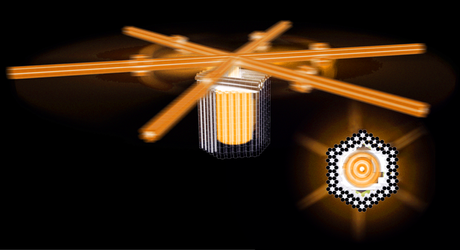
Precision design needs precision validation. Our lab has direct access to high-end transmission electron cryo microscopy equipment, consisting of a 300kV FEI Titan Krios with direct electron detector and two 120kV-class TEM for sample screening purposes. For image processing we have access to a high-performance computing cluster. We use iterative cycles of design and structural analysis to create components and devices with ever increasing accuracy. The goal is to hone our design skills and ultimately learn how to build complex molecular structures with atomic accuracy. These capabilities appear essential to rationally realize molecular recognition and enzymatic catalysis tasks.
The image on the left shows a slice through a multi-layer DNA-based nanostructure, revealing the hallmark signatures of double-helices including major and minor grooves, and also revealing covalent phosphate backbone connections that are used to stabilize the structure internally.